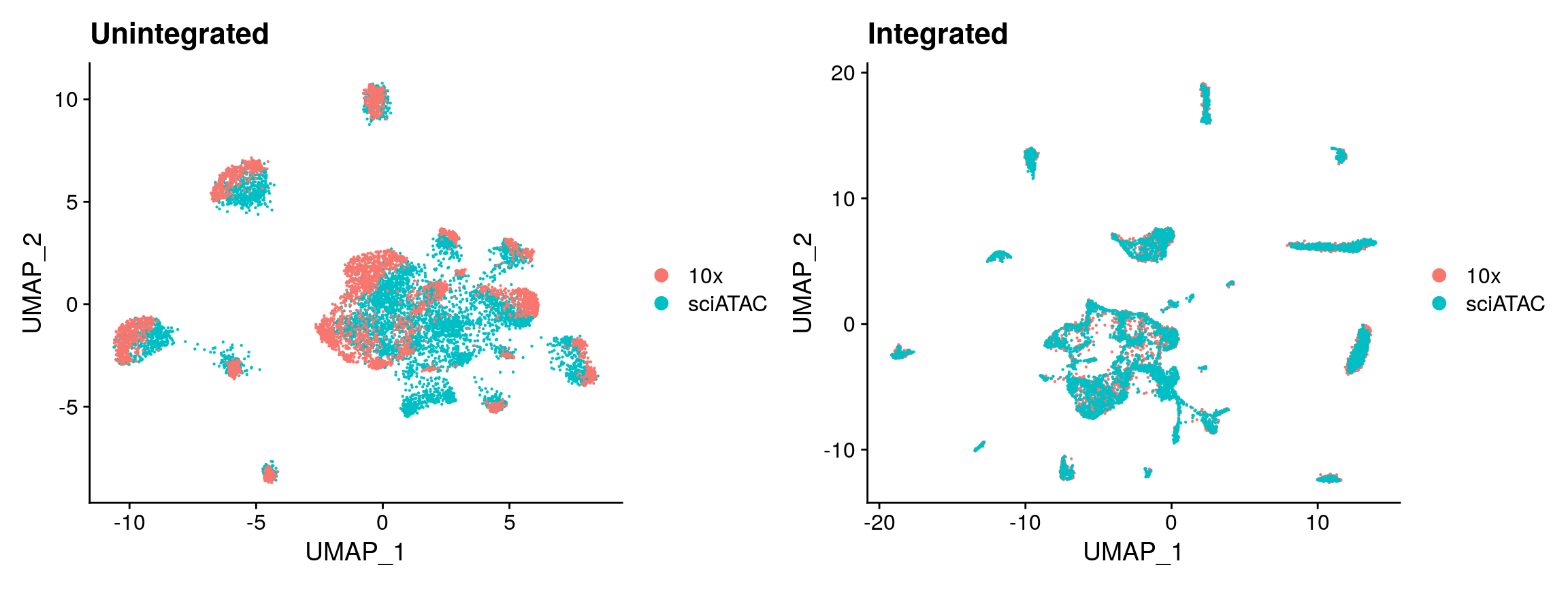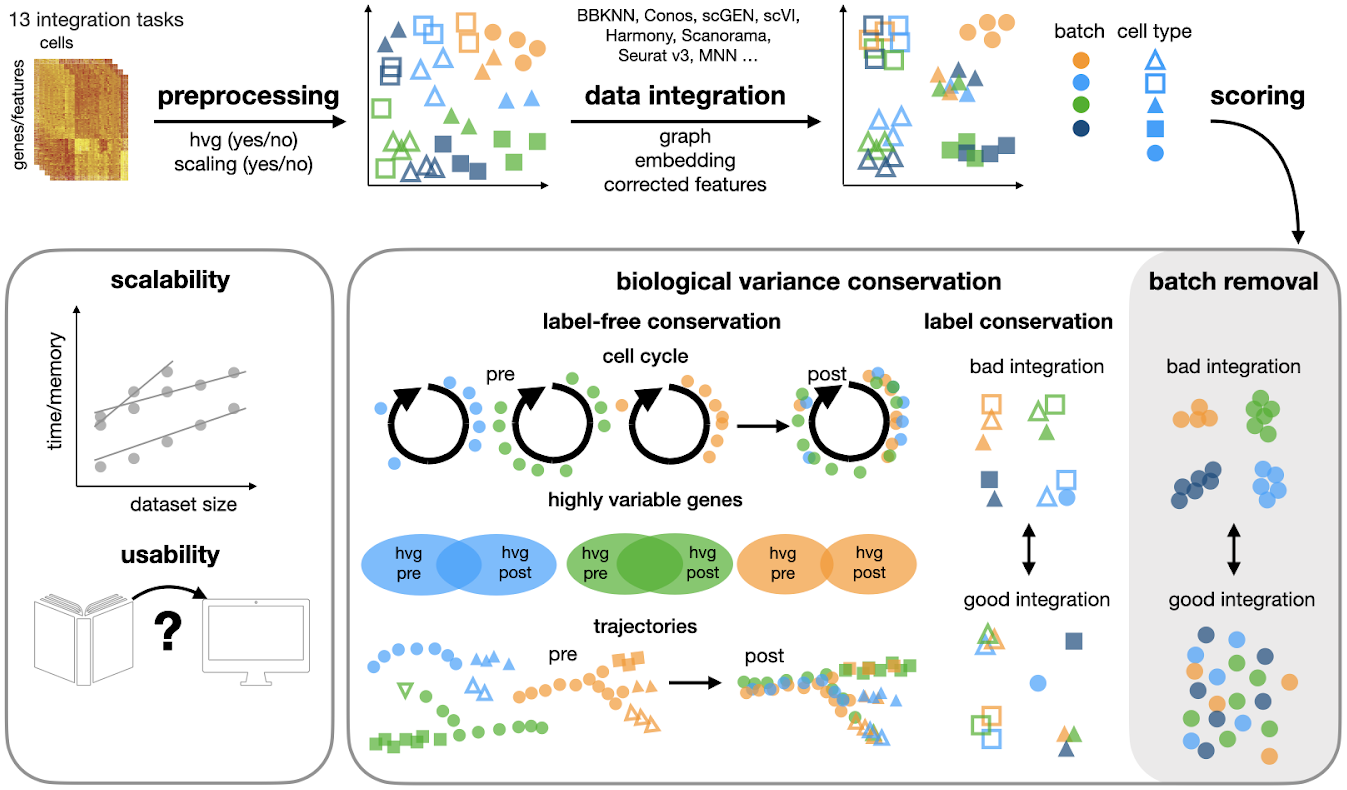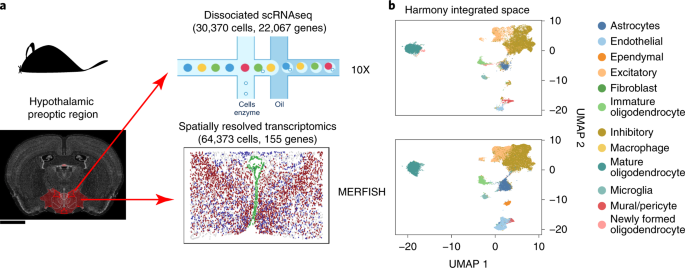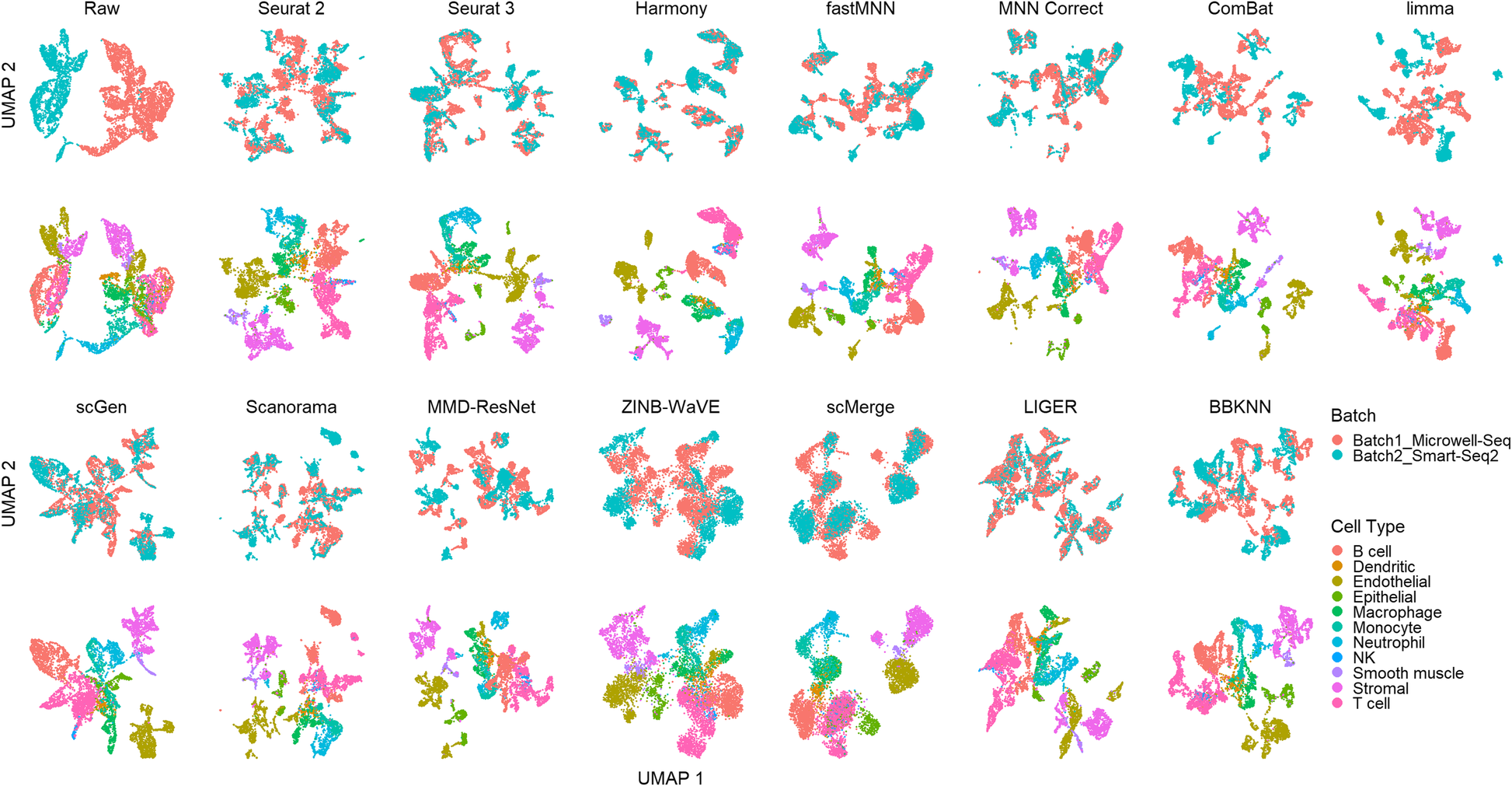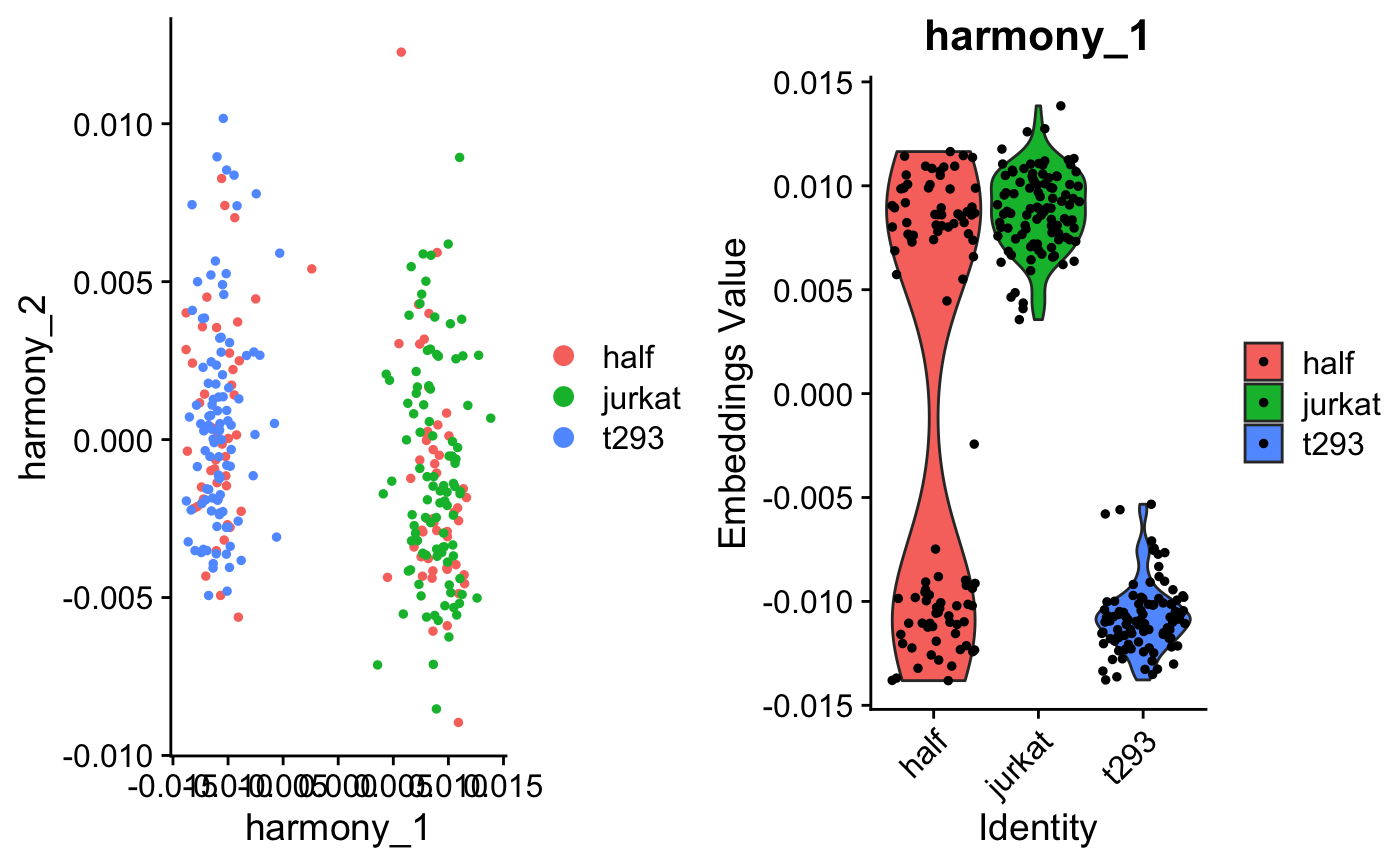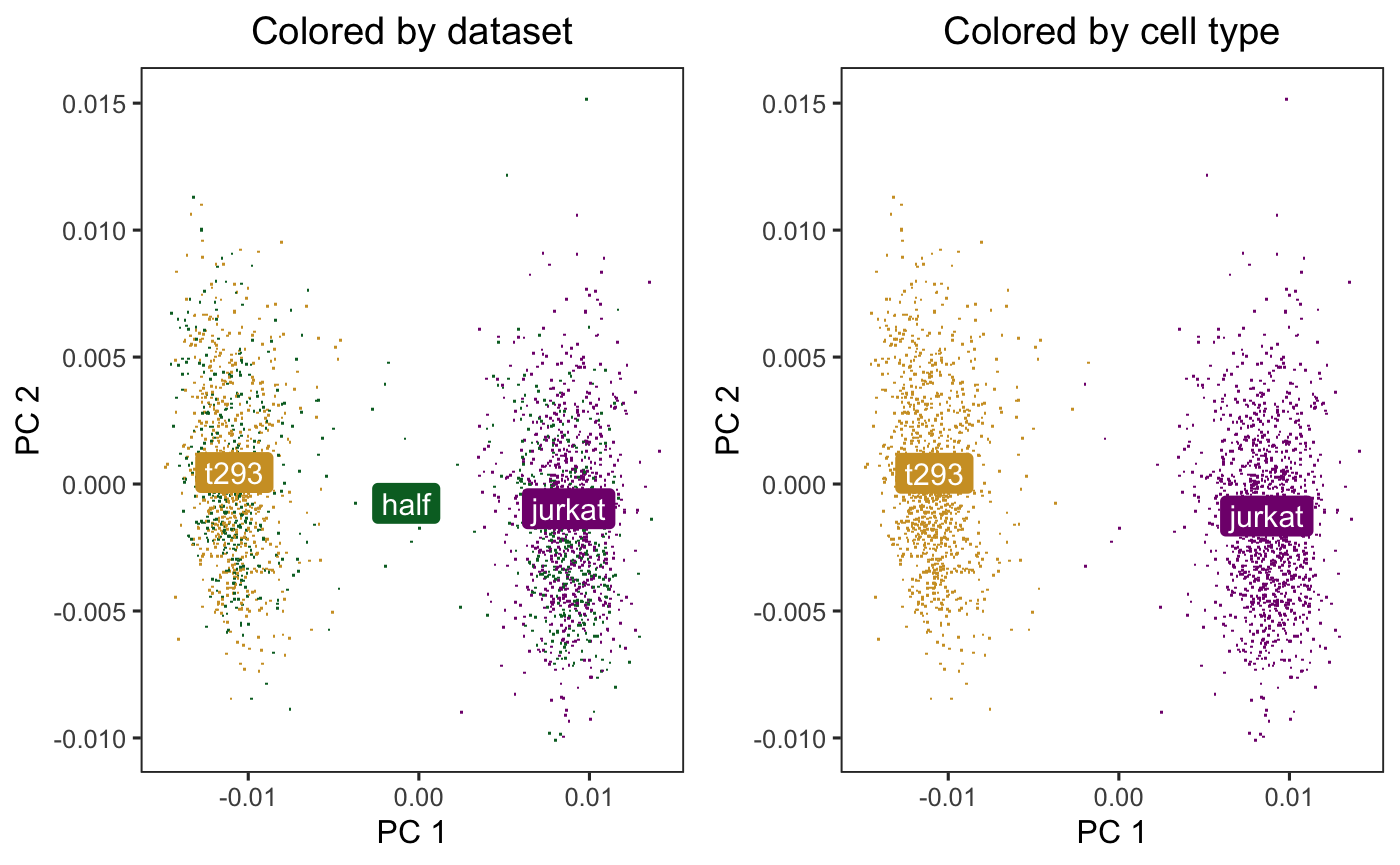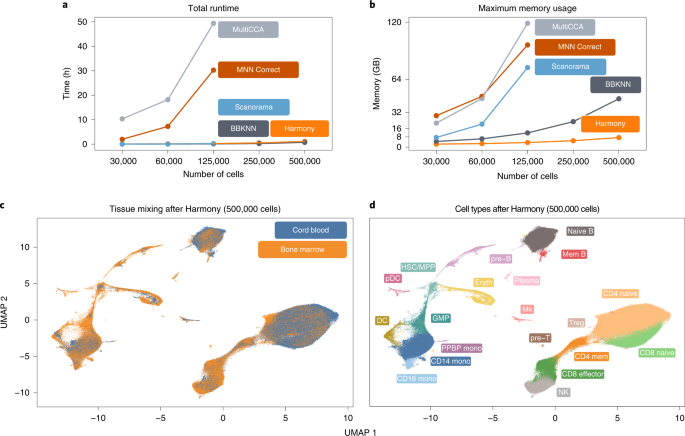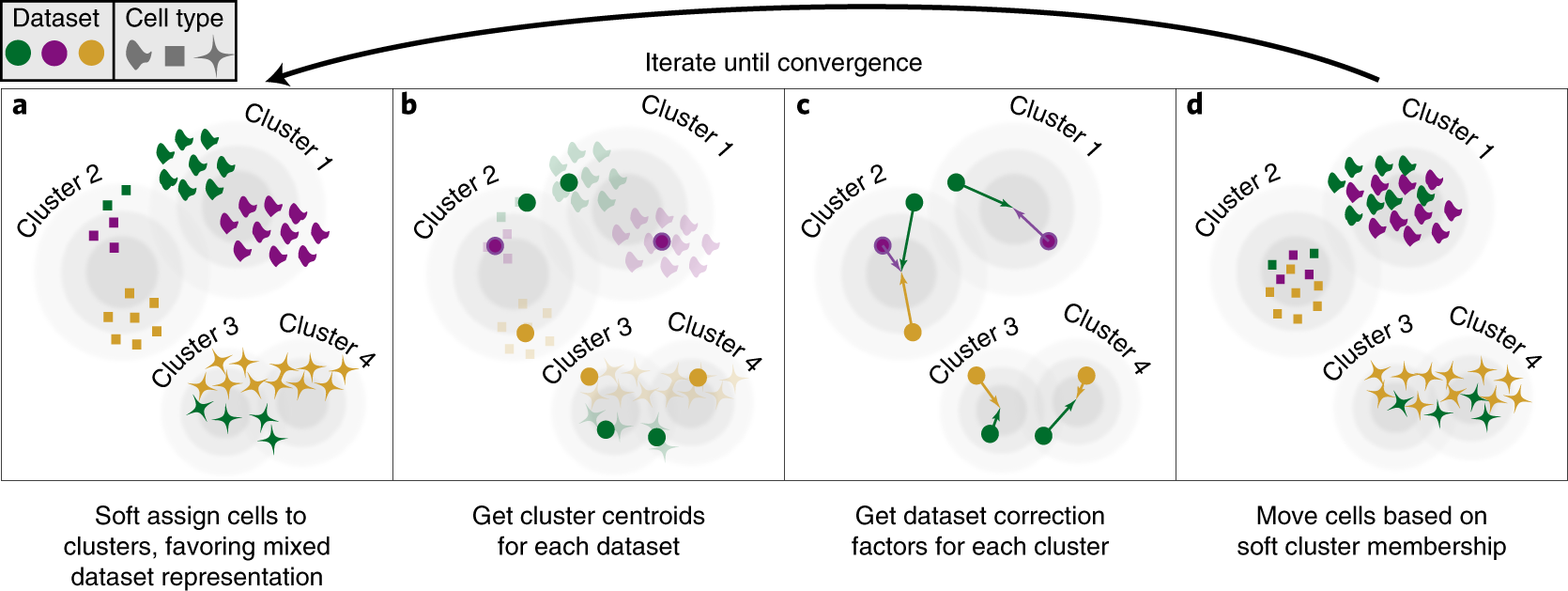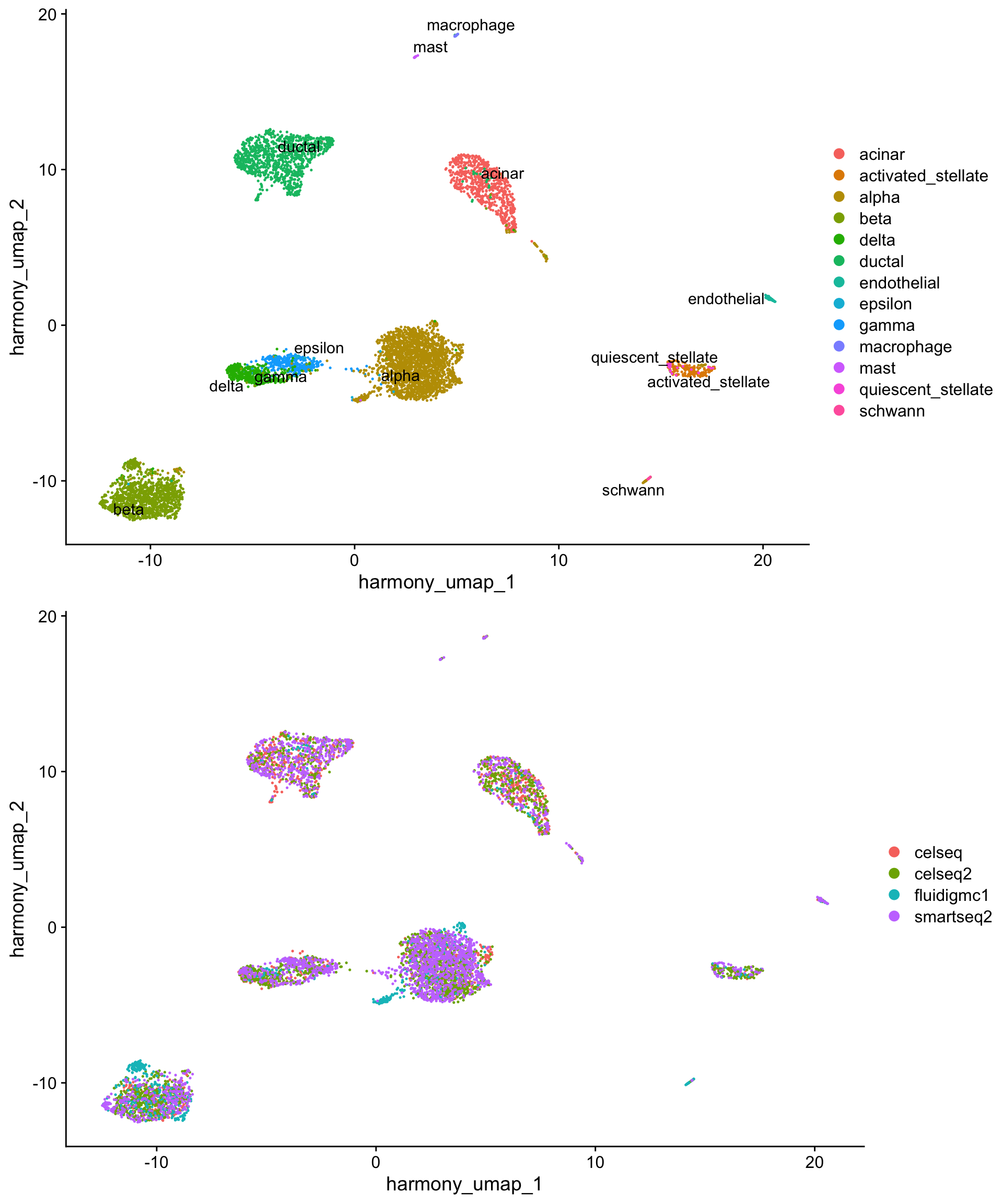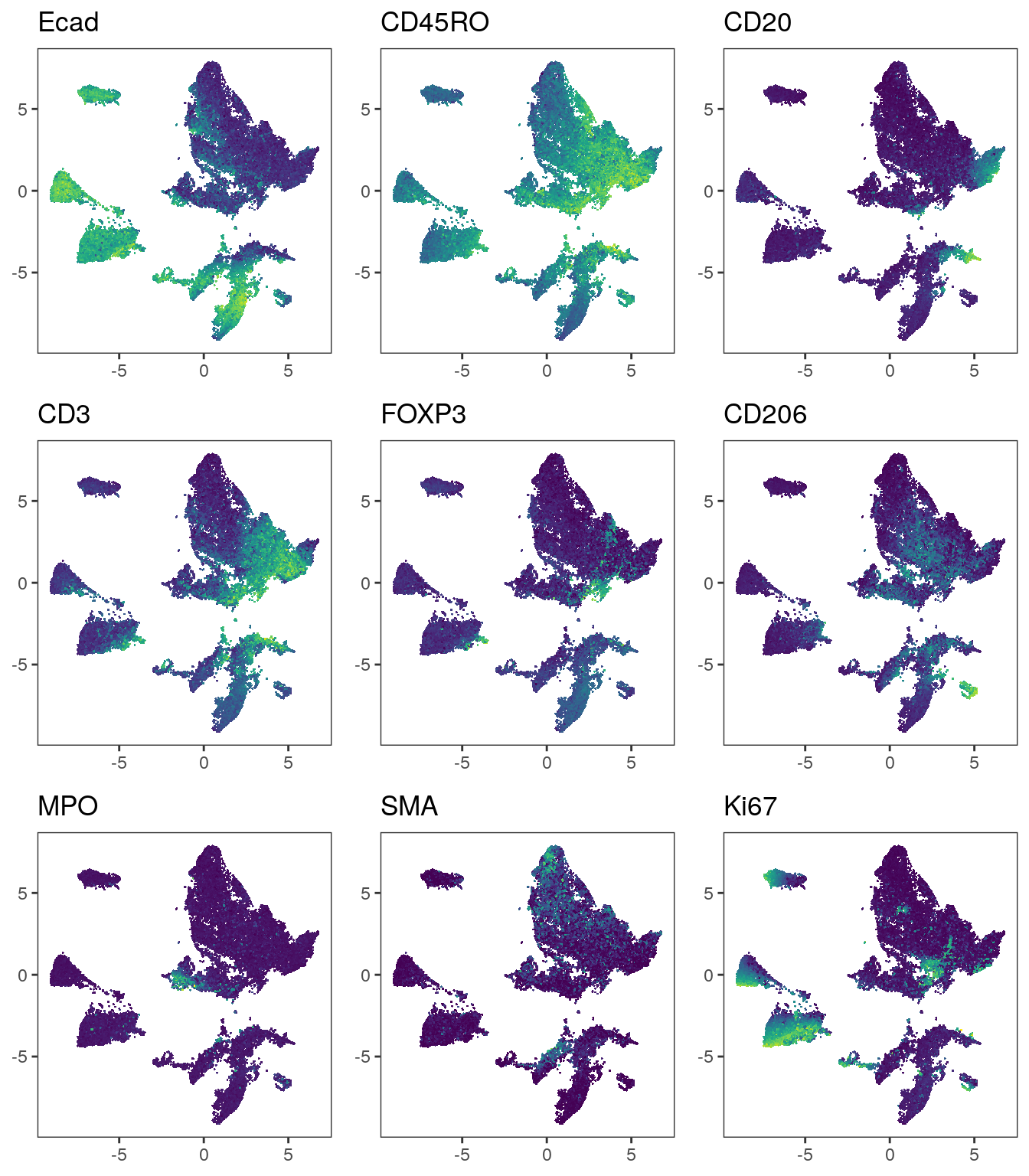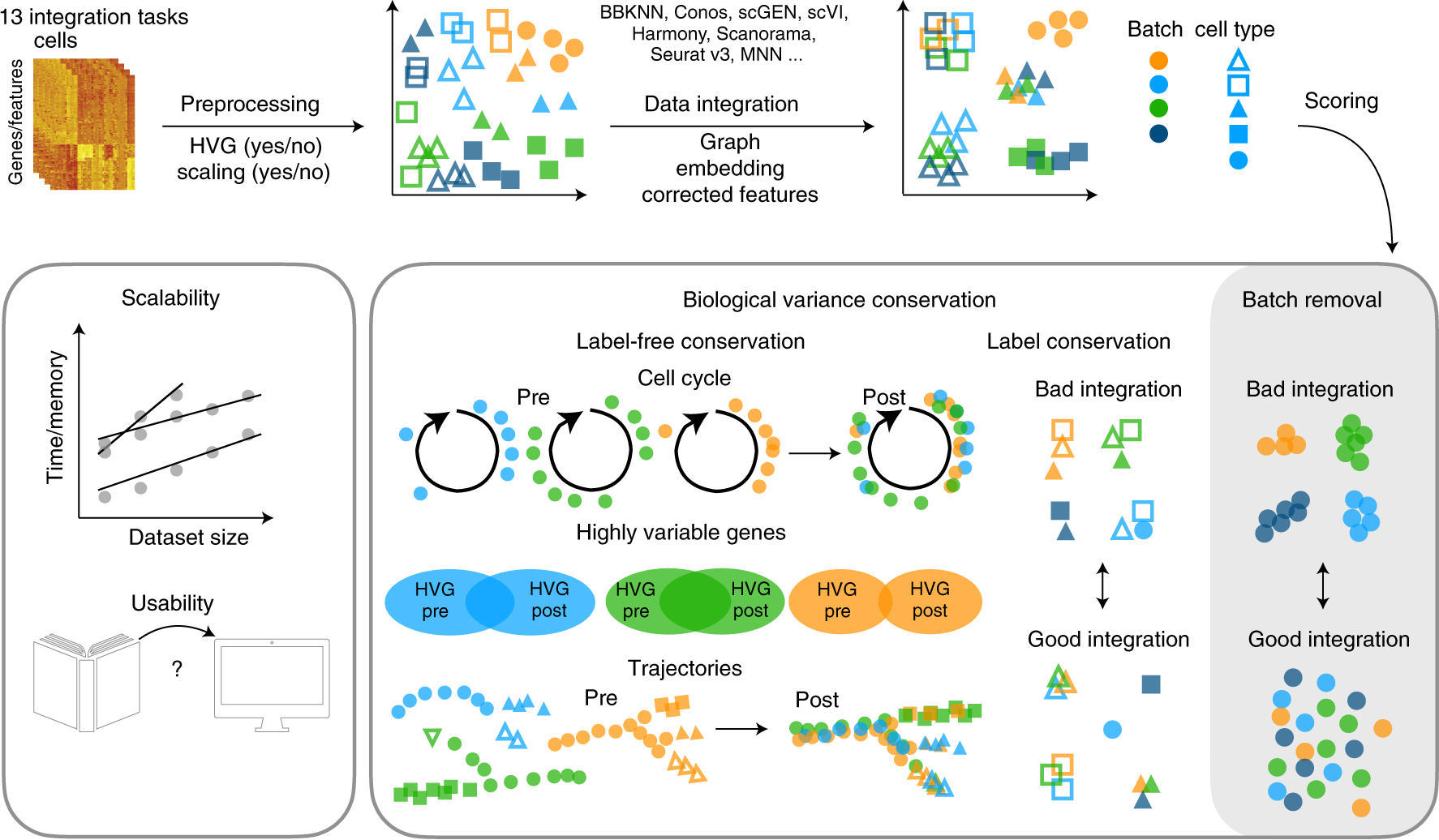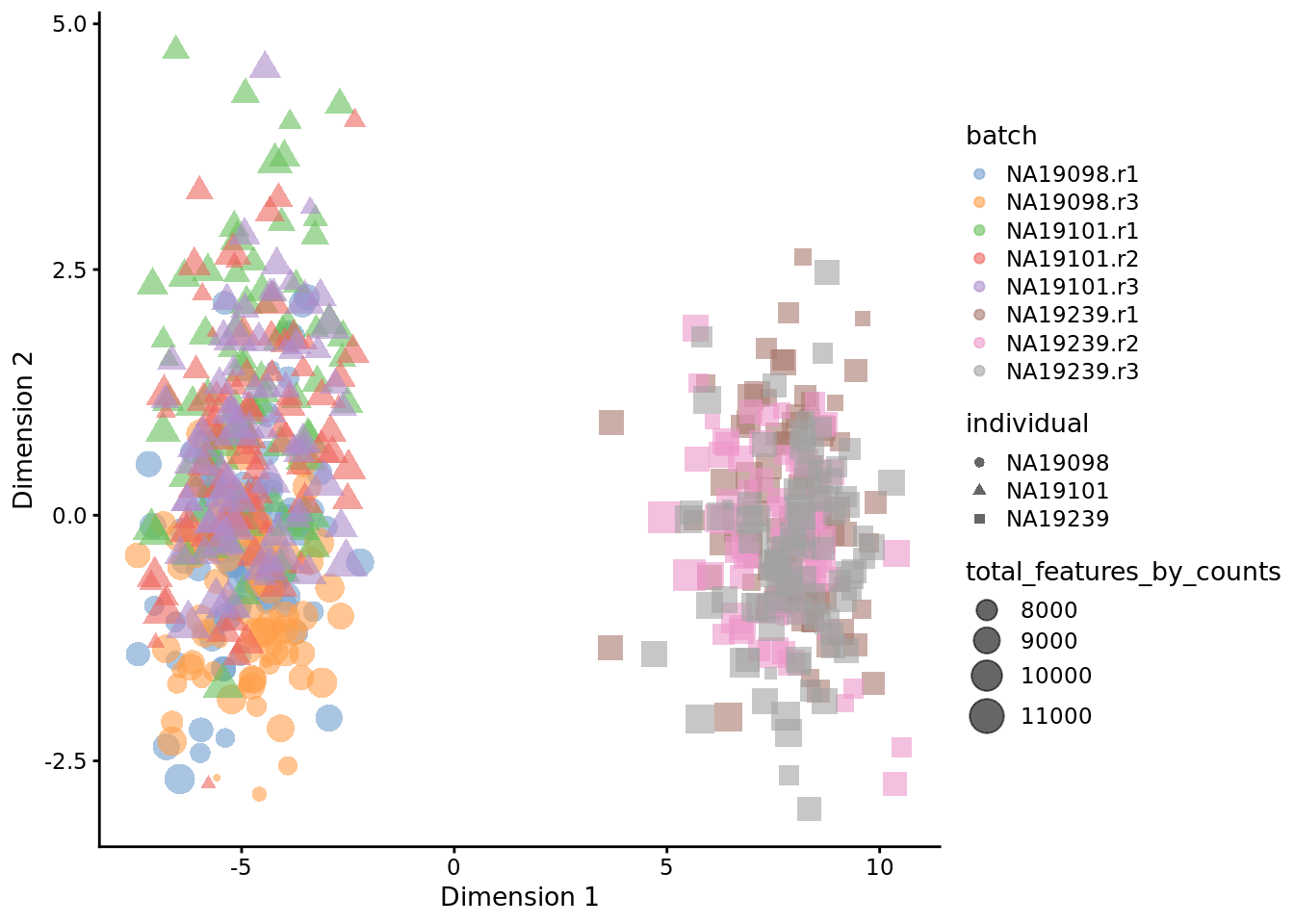
A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text
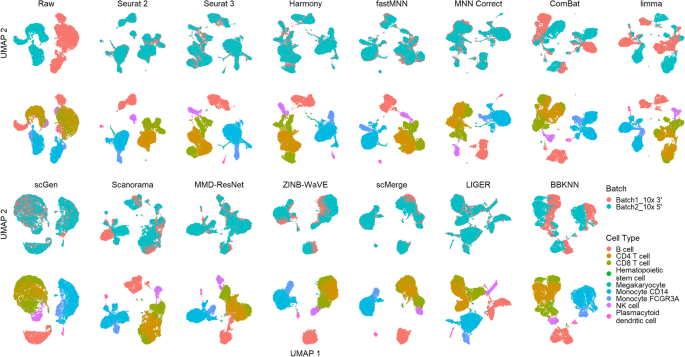
A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text

A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text
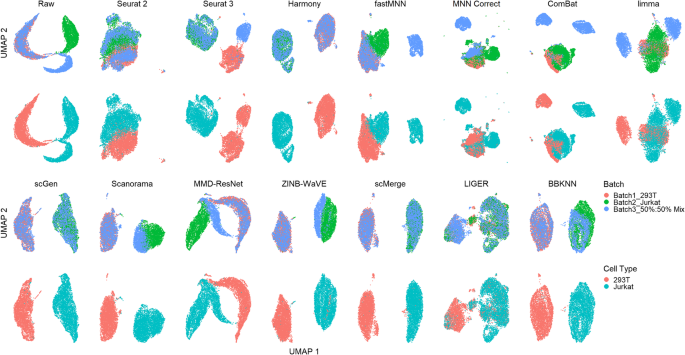
A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text

A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text